Novel introductions and epidemic dynamics of the sudden oak death pathogen Phytophthora ramorum in Oregon forests
Sudden oak death caused by Phytophthora ramorum has been actively managed in Oregon since the early 2000’s. To date, this epidemic has been driven mostly by the NA1 clonal lineage of P. ramorum, but an outbreak of the EU1 lineage has recently emerged. Here we contrast the population dynamics of the NA1 outbreak first reported in 2001 to the outbreak of the EU1 lineage first detected in 2015. Diversity of EU1 was very low and isolates were spatially clustered (< 8 km apart), suggesting a single EU1 introduction. In contrast, the older NA1 populations were more polymorphic and spread over 30 km2. Principal component analysis supported two to four independent NA1 introductions. The NA1 and EU1 epidemics infest the same area but show disparate demographics owing to initial introductions of the lineages spaced 10 years apart. Comparing these epidemics provides novel insights into patterns of emergence of clonal pathogens in forest ecosystems.
Citation: *Carleson, N.C., *Daniels, H.A., Reeser, P.W., Kanaskie, A., Navarro, S.M., LeBoldus, J.M., Grünwald, N.J. 2020. Novel Introductions and epidemic dynamics of the sudden oak death pathogen Phytophthora ramorum in Oregon forests. Phytopathology.
Full text available soon!
From genomes to forest management – tackling invasive Phytophthora species in the era of genomics
Species of Phytophthora pose one of the most serious biosecurity threats to forest ecosystems worldwide. Despite management efforts and increased awareness of forest pathogens, there is continued introduction and spread of Phytophthora species. Uncertainty about the center of origin for many of the invasive species hampers disease control efforts. Additionally, the management efforts are often made impossible either by the vast host range or the extreme susceptibility of naïve hosts. In this review, we discuss how genomics has shed light on the extent of spread and destruction caused by invasive Phytophthora species, and how approaches leveraged by genomics can be applied to enhance the management of these invasive forest pathogens.
Citation: Keriö, S., Daniels, H.A., Gómez-Gollego, M., Tabima, J.F., Lenz, R.R., Søndreli, K.L., Grünwald, N.J., Williams, N., McDougal, R., LeBoldus, J.M. 2019. From genomes to forest management – tackling invasive Phytophthora species in the era of genomics. Canadian Journal of Plant Pathology 42:1, 1-29.
Full text available upon request (contact me!).
Doctoral thesis, Oregon State University, Spring 2021
Advisor: Jared LeBoldus, PhD
Sudden Oak Death, a disease of unknown origin, was found infecting the coastal forests of California in 1995. Around the same time, researchers in Europe discovered a twig blight infecting ornamental plants was caused by oomycete species Phytophthora ramorum. Through conversations between these two groups, Sudden Oak Death was linked with the pathogen Phytophthora ramorum. It was discovered in the forests of Oregon in 2001, and in Europe and the UK around the same time. In Britain, the name Sudden Larch Death is used to refer to P. ramorum infections found in plantations of exotic Larix kaempferi (Japanese larch).
Phytophthora ramorum is spread aerially by wind and wind-driven rain, and it survives in soil, leaf litter, and water. It is spread among forest canopies, and especially in nurseries. There are more than 100 host species, most of which are foliar hosts on which sporangia germinate or release zoospores. Bark hosts, also called bole hosts, become infected when xylem tissues are colonized, and include various oaks (Quercus spp.), tanoak (Notholithocarpus densiflorus), larch (Larix spp.), and beech (Fagus spp.). Symptoms of Sudden Oak Death vary among species, but include bleeding bole cankers, cambial lesions, wilting shoots, rapid browning and subsequent death of bole or bark hosts. Understory host species frequently show nonlethal foliar symptoms including necrotic lesions, shoot dieback, and vascular discoloration. Much effort has been spent increasing the speed and accuracy of assays to differentiate between Phytophthora species and between the various lineages of P. ramorum. Despite these early detection efforts, the disease is common in nurseries, and has been transported globally through nursery trade. It is a federally regulated pathogen in the United States (APHIS, 2007), yet infected plants were recently shipped to 18 states in the eastern U.S. The high inoculum levels in nurseries has led to significantly higher genotypic diversity in nursery populations than in forest populations. Should P. ramorum escape from a nursery in the eastern U.S., climatic variables are suitable for establishment of the pathogen, and there are a number of native species at high risk for infection.
My dissertation is organized into three sections, the first of which is condensed into the poster below.
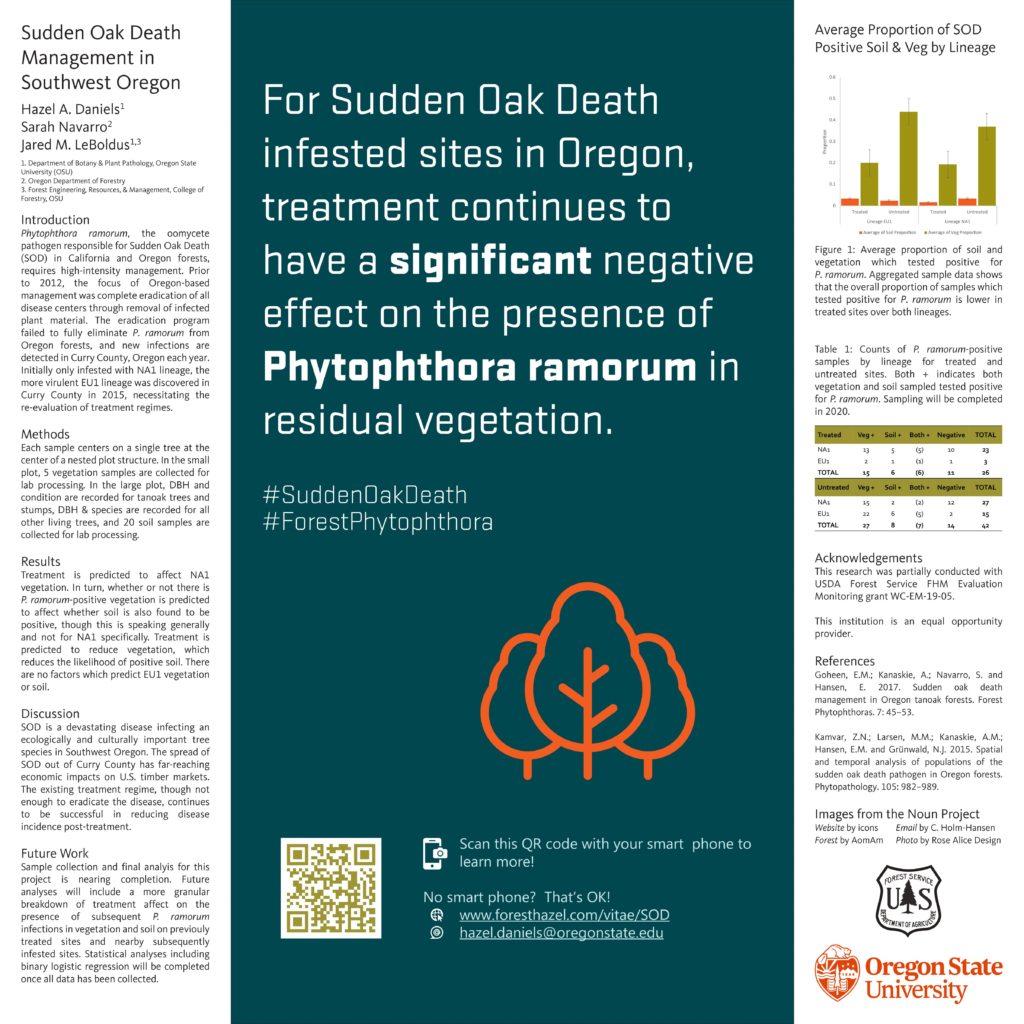
AIM 1 – Determine the efficacy of current management practices by detecting the presence or absence of P. ramorum inoculum in understory vegetation and soil from treated and untreated sites which have previously tested positive for P. ramorum.
In order to achieve the stated aim, ca. 100 sites will be visited over three years. Sampled sites were selected from known treated or untreated infested sites. A small number of uninfested sites within the QZ were also selected. Untreated infested sites, especially those infested with the EU1 lineage, will be resampled after treatment when possible. The expected outcome from this research is an understanding of the efficacy of management practices on the spread of Phytophthora ramorum in Oregon. Based on current understanding of Sudden Oak Death, and the stated goal of the Sudden Oak Death task force (e.g. slow the spread of SOD in Oregon) the following hypotheses have been put forth:
- Current treatment regimes used by Oregon Sudden Oak Death task force (including herbicide treatment, cutting, and burning) are effective in reducing the amount of Phytophthora ramorum inoculum in soil at previously treated sites.
- Current treatment regimes are effective in reducing the amount of Phytophthora ramorum inoculum in understory vegetation at previously treated sites.
- There will be higher rates of isolation from soil and vegetation at EU1 infested sites. Increased sporulation of EU1 will lead to greater reservoirs of inoculum at these sites.
Preliminary results: Two years of sampling have been completed. Once the third and final sampling season is complete, the full data set will be analyzed. A breakdown of sampled sites and positive vegetation and soil samples for 2018 and 2019 shows 68 total sites sampled over the two-year period. Of these, 50 sites were infested with NA1 and 18 were infested with EU1. Thirty six of these 68 sites were untreated (21 NA1 and 15 EU1), and the remaining 32 sites were treated (29 NA1 and 3 EU1). Two uninfested sites were sampled in 2019 but are not included in current analysis.
When data is aggregated and viewed as a proportion of the total number of samples within each group (lineage, treatment status, sampled material), the results of treatment on vegetation for both NA1 and EU1 lineages are clear. There were very few positive soil samples overall, which affected our ability to perform analyses on this subset of our data.
Binary logistic regression was performed on the combined EU1/NA1 dataset, separated by sample material (soil or vegetation). For combined vegetation data, any treatment changed the log odds of P. ramorum-positive vegetation by -2.1. Treated sites had a negative association with infected vegetation, which aligns with Hansen et al. (2019). For combined soil data, the presence of P. ramorum-positive vegetation changed the log odds of there being positive soil samples by 2.42. A site with P. ramorum-infected vegetation is more likely to have positive soil samples than a site without positive vegetation samples. For both soil and vegetation models, it is plausible that the data emanate from a logistic model that includes these variables. We can reject the null hypothesis that the deviance of the model with these variables is the same as the deviance of the model with only the constant. The final sampling season will focus exclusively on collecting samples from sites which tested positive for the EU1 lineage of P. ramorum in order to strengthen these analyses and allow for separate NA1 and EU1 analysis.

AIM 2 – Reveal genetic and geographic relationships between P. ramorum isolates of each lineage (NA1, EU1) over a period of four years using whole-genome sequencing.
Since the discovery of Sudden Oak Death in Oregon, isolates collected from infected tanoak trees have been maintained and kept in long-term storage. Samples dating back to the first infections in 2001 are still available for study. It is important to assess and reassess the efficacy of management in the face of increasing numbers of infections, changes in infested areas, and northward movement of the pathogen (Figure 4). This is an unprecedented opportunity in Sudden Oak Death research, as the disease has been monitored and treated since its introduction to Curry county. Using the existing isolate collection, we can test hypotheses that can inform the future. Therefore, the expected outcome from this study is an expanded understanding of the epidemiology of the P. ramorum outbreak in Curry county, with specific attention given to the newly introduced EU1 lineage. With this expected outcome in mind, the following specific hypotheses are presented:
- Subsequent Phytophthora ramorum infections in trees will occur geospatially closer to trees which were previously infected than expected by chance.
- Subsequent Phytophthora ramorum infections in trees geospatially near to recently treated infected trees will be more closely genetically related to those initially treated infected trees than ramorum infections in trees geospatially further away.

AIM 3 – Determine the suitability of Radiata pine (Pinus radiata) as a bole or seedling host for P. ramorum.
Radiata pine is one of the most important plantation species in New Zealand forest industry. Previous research by Hüberli et al. in 2008 showed that Radiata pine branches had large lesions and branch girdling after inoculation, and foliage became symptomatic after inoculation. Thus, there is a need to further explore the susceptibility of Radiata pine to P. ramorum. This project focuses on logs in a forestry context, which are larger in diameter than those of Hüberli’s work. In addition, the long list of host species susceptible to P. ramorum has not been exhaustively covered. Thus, if the pathogen is discovered in New Zealand, there is a potential risk that P. ramorum could establish a new disease system with Radiata pine as a host. This project was initiated to determine the risk, if any, to Radiata pine logs in the context of forest harvest. The following research questions were posed:
- Radiata pine logs will not be susceptible to P. ramorum infection under field conditions or through artificial inoculation. We will be able to isolate the pathogen from artificially inoculated lesions, but not through lesions found on the field inoculated logs.
- Radiata pine seedlings will be susceptible to P. ramorum infection under field conditions and through artificial inoculation, and we will be able to isolate the pathogen from any lesions generated through this experiment.